***
The field of bioinformatics and computational biology is a rapidly emerging area with the potential to advance scientific discovery to the degree required to solve many of the challenging biological problems of the 21st century. WSU has a rich history of fostering the kind of interdisciplinary science that is needed in such an endeavor. WSU computer scientists, in collaboration with leading plant scientists and microbiologists both at WSU and world-wide, are working on projects that include genome discovery for economically important plant crops, vaccine development, decoding the evolutionary history of microbes, and understanding the functional basis for genes and proteins involved in bioenergy and renewables. Recent and ongoing projects include:
- Designing innovative algorithmic solutions for data-intensive life sciences applications. Applications include genome assembly and annotation for economically important plant crops, identification of proteins involved in bioenergy, and decoding gene regulatory and protein-protein interaction networks.
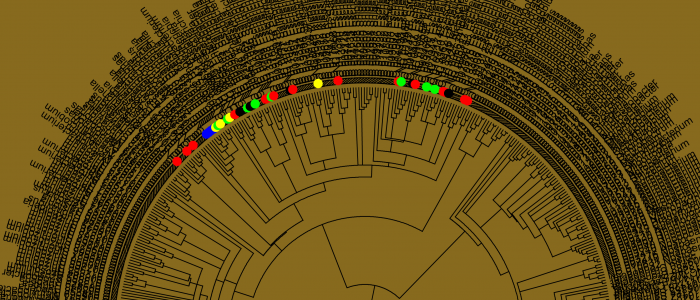
- Developing models and algorithms for microbiology applications including microbial evolution and phylogenomics, epidemiology, vaccine development, and bacterial source tracking.
- Developing machine learning software programs to predict antibiotic resistance genes in bacterial pathogens and to predict type IV secretion system effector proteins in both bacterial and apicomplexan pathogens.
- Developing scalable parallel algorithms for data-intensive biological applications using next-generation supercomputers. Target parallel architectures include massively parallel distributed and shared memory supercomputers, cloud computing platforms, and multicore hardware accelerators.
- Developing algorithms for protein and metabolite identification in complex mixtures by high-throughput mass spectrometry.
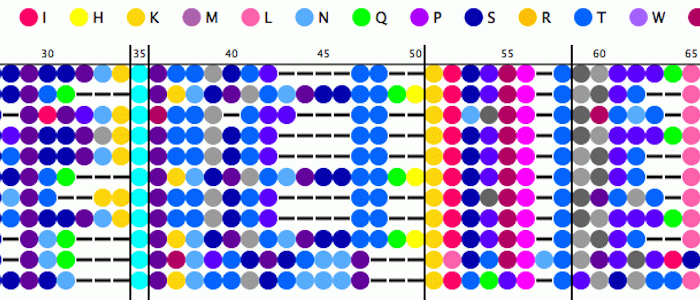
- Developing robust software tools for management and analysis of short sequence repeat data on pathogens. One of our current such tools, RepeatAnalyzer, has functionalities for storing, managing, searching and identifying repeats in a dataset on the bacteria Anaplasma marginale.
- Developing efficient sequence similarity network (SSN) models to enable scientific discovery. Our recent work on the Directed Weighted All Nearest Neighbors (DiWANN) network model for sequence data provided new insight into genotype distribution of a pathogen. A short (2 min) video that highlights some of the mathematics underlying DiWANN and the science it enabled is available here: https://www.youtube.com/watch?v=bOwxDwVE2oc&feature=youtu.be
- Research funding from the National Science Foundation, National Institutes of Health, Department of Agriculture, and Department of Energy.